What’s new in SAMSON 2020 R3
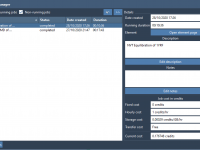
The new SAMSON 2020 R3 release brings numerous new features and improvements throughout the core of SAMSON, so let’s dive right in! Cloud computing One of the most exciting features of SAMSON 2020 R3 is the introduction of cloud computing capabilities. In many design situations, a personal computer might have too little processing power to perform advanced calculations (e.g. high-throughput screening, some molecular dynamics simulations, etc.). In agreement with our vision of democratizing molecular design, we are introducing the possibility to…