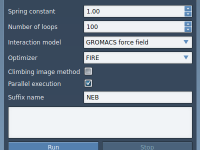
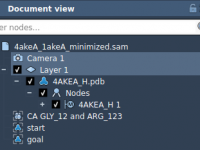
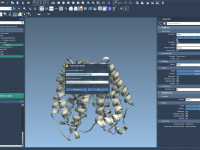
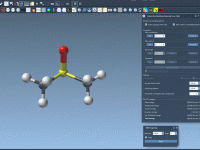
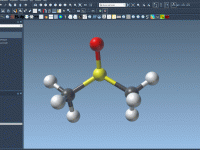
Export atom trajectories along paths
In this tutorial, we will show how to export atoms positions (for all atoms in the system or for a selection of atoms) along paths in PDB files thanks to the Export Along Paths app. This could be useful for example if you want to generate reaction coordinates for free energy calculations. Requirements Export Along Paths Extension Before starting the tutorial, please download the 1pv7_ligand_complex_with_paths (~13 MB) which contains files for this tutorial. Tutorial Launch SAMSON. Load 1pv7_ligand_complex_with_paths.sam file…