SAMSON has a signals and slots mechanism on which SAMSON Extensions may rely to develop adaptive and incremental algorithms, process events (e.g. structural, visual events, etc). There are many different events emitted in SAMSON, please, refer to the Signals and slots section for more information.
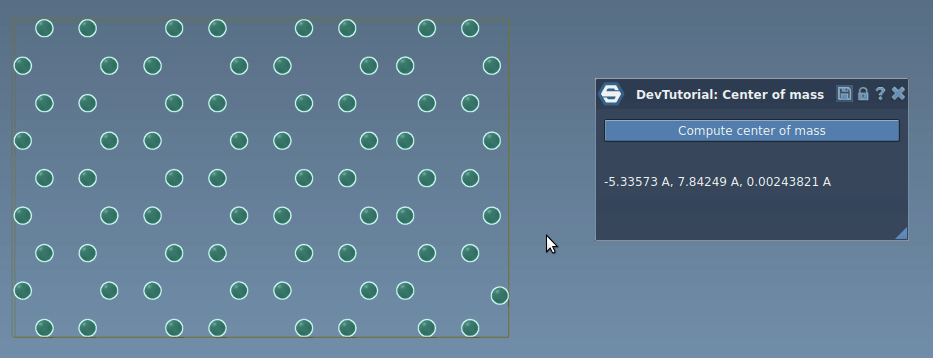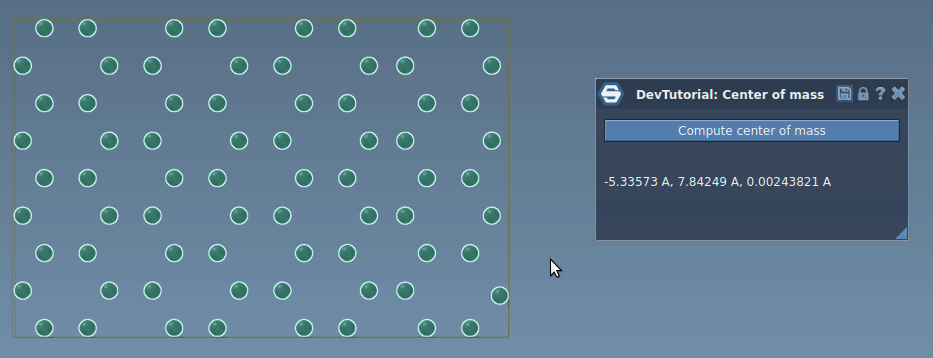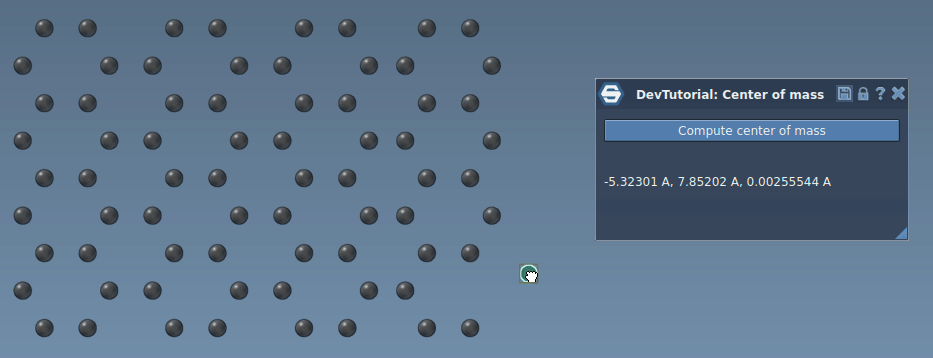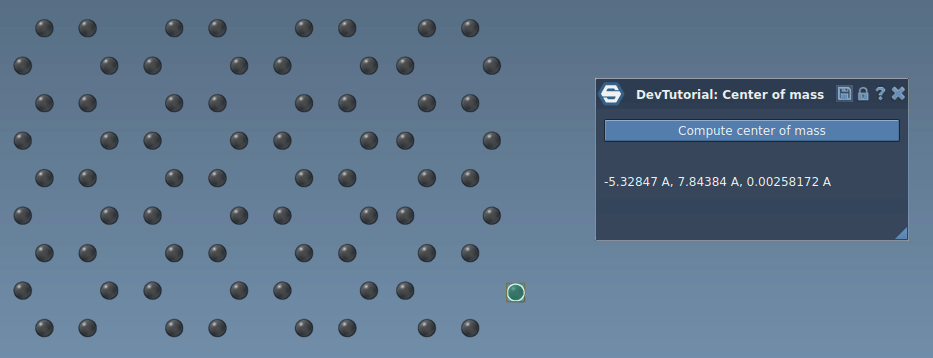
In this tutorial, you will learn how to process signals in SAMSON on an example of an App that incrementaly computes the center of mass of the system when atoms move. Re-computing the center of mass of the whole system when just a few atoms moved might be computationally heavy and unnecessary, we can rather update the center of mass incrementally based only on atoms that moved by modifying their contribution into the center of mass. When an atom is moved via e.g. the point editor it emits the SBStructuralEvent::AtomPositionChanged event, it is this event which we will be using in this tutorial to incrementally update the center of mass.
The source code for an Extension created in this tutorial can be found at https://github.com/1A-OneAngstrom/SAMSON-Developer-Tutorials.
Generating an Extension
We will start by creating a new SAMSON Extension called CenterOfMass thanks to the SAMSON Extension generator (please, refer to the SAMSON Extension generator tutorial for a reminder on how to use it):
- launch SAMSON and run the SAMSON Extension generator;
- specify the path to a folder where you want to develop your SAMSON Extensions;
- name the SAMSON Extension as CenterOfMass and add some description;
- add an App class (called
SECenterOfMassApp); - generate the SAMSON Extension.
The SAMSON Extension generator generates an app class that implements the functionality of the app and is derived from the SBDApp class, and a GUI class that implements the user interface of the app and is derived from the SBGApp class.
Setting the description of the Extension
Open the header file of the app descriptor (file: SECenterOfMassAppDescriptor.hpp) and modify the class description:
Open the source file of the app GUI (file: SECenterOfMassAppGUI.cpp) and modify the getName function to have a user-friendly description of the app inside SAMSON:
Now, try building the Extension and when you launch SAMSON you should be able to find it in Home > Apps. For now, it has no interface and does nothing, let's now add functionality to it.
Setting up the interface
Let's start by adding the interface. Open the SEAtomShakerAppGUI.ui file. Clear the text in the label and add a button to the user interface named pushButtonComputeCenterOfMass.
Add the following slot: onComputeCenterOfMass()
and connect the clicked() signal from the button to the added slot:

Computing the center of mass
Let's first implement a simple computation of the center of mass of the selected atoms.
Open the header file of the GUI class (class: SECenterOfMassAppGUI, file: SECenterOfMassAppGUI.hpp) and include the SBVector3.hpp header file (necessary for SBPosition3):
Add to the GUI class the onComputeCenterOfMass() public slot and a function to update the label with the new center of mass:
Implement the slot function onComputeCenterOfMass() as follows:
Here, we invoke the app's function for computing the center of mass (we will implement this function later in the section). Even for simple apps, it is preferable to separate the user interface from the core functionality of the app.
Implement the updateCenterOfMass() function to show the center of mass in the interface:
Let's now implement the computation of the center of mass in the app class. Open the header file of the app class (SECenterOfMassApp.hpp) and include SBAtom.hpp and SAMSON.hpp headers:
And add the following function and variable in the app class:
For now, we implement the computeCenterOfMass function without taking into account moving atoms:
Now, try building the Extension and when you launch SAMSON you should be able to find it in the App menu or in the App toolbar. Load or create a molecule, select it, and compute the center of mass.
Processing signals
Let's now implement the computation of the center of mass in the incremental way thanks to the signals and slots mechanism. For that, we connect structural signal of each atom in the selected system to a slot, in which we will be listening to the SBStructuralEvent::AtomPositionChanged event emitted by an atom when its position has changed.
And add the following function and variables in the app class (SECenterOfMassApp.hpp):
The atomIndexer will be used for storing indices of atoms for which the center of mass should be computed and the positionArray will be used for keeping the previous positions of atoms.
Modify the constructor and destructor of the app class (SECenterOfMassApp.cpp) as follows:
Implement the function for processing the structural event as follows:
Here, we update the center of mass based on an event emitted when the particle position has changed. Note that for performace reasons this event is not emitted if the particle position was changed during the simulation or through some of the editors.
First, we get an index in the atomIndexer of an atom that emitted the event, we substruct this atom's old position from the center of mass and add its new position. Then we remember the current position of the atom and we request the app's interface to update the label.
We want the following behavoir from the app: once the button in the interface is clicked, the center of mass should be computed for the currently selected system and later it is updated if any atom from the selected system changes its position. For that, we modify the implementation of the computation of the center of mass as follows:
Here, we first clean the array with old positions of atoms, and disconnect previously added atoms from the slot. Then we find atoms in the selected system, add them into the atomIndexer, and connect them to the slot. Finally, we compute the center of mass and update it in the interface of the app.
Now, you have a simple implementation of an incremental computation of the center of mass. Build the Extension and launch SAMSON. Load or create any molecule, select it, click on "Compute center of mass" button in the app, and try moving an atom. You should see that that the center of mass is updated in the app interface.