In this tutorial, we will create a simple spring model force field such that a virtual spring links two pairs of atoms connected by a covalent bond.
In SAMSON, a force field is presented as an interaction model.
The source code for an Extension created in this tutorial can be found at https://github.com/1A-OneAngstrom/SAMSON-Developer-Tutorials.
Generating an Extension
We will start by creating a new SAMSON Extension called Springs thanks to the SAMSON Extension generator (please, refer to the SAMSON Extension generator tutorial for a reminder on how to use it):
- launch SAMSON and run the SAMSON Extension generator;
- specify the path to a folder where you want to develop your SAMSON Extensions;
- name the SAMSON Extension as Springs and add some description;
- add an Interaction Model class (called
SESpringsInteractionModel); - generate the SAMSON Extension.
The SAMSON Extension generator generates an interaction model class (derived from the SBMInteractionModelParticleSystem class) and a property widget class for the interaction model.
Setting the description of the Extension
Open the header file of your interaction model descriptor (SESpringsInteractionModelDescriptor.hpp) and modify the class description:
Do the same for the property window descriptor (SESpringsInteractionModelPropertiesDescriptor.hpp). And modify the getName function of the property window (SESpringsInteractionModelProperties.cpp) to have a user-friendly description of your interaction model inside SAMSON:
Now, try building the SAMSON Extension.
Start SAMSON and open any file containing atoms or add atoms in the data graph, then create a new simulator (Simulation > Add simulator or ctrl / Cmd⌘ + shift + M ): your new Extension should be in the list of the available interaction models.
Initializing the interaction model
Open the header file of your interaction model (SESpringsInteractionModel.hpp) and include SBBond.hpp.
Add the following functions and variables in the SESpringsInteractionModel class that will be used in the force field.
Here:
initializeBondSpringsModelis a function that initializes the bond spring interaction model;updateBondSpringsInteractionsis a function that updates the bond spring interactions;particleIndexeris an indexer that will point to the atoms (structural particles) of your system;bondSpringStiffnessregulates the stiffness of bond springs;springLengthVectorstores the equilibrium length for the springs of your model;springBondVectorstores the bonds of the springs of your model.
In both constructors of the interaction model (SESpringsInteractionModel.cpp) initialize the bondSpringStiffness variable to 1.0:
Modify the initializeInteractions (SESpringsInteractionModel.cpp). This function is called to set your interaction model up and initialize the energy and forces.
Here, we invoke the initialization function for the bond spring interaction model and emit the changed signal to let SAMSON know that the energy and forces have been changed.
Implement the initializeBondSpringsModel function as follows:
First, we clear the vectors used to store the bonds and their equilibrium lengths. Then we initialize the particle indexer (particleIndexer) based on the particle system (particleSystem) the interaction model is attached to. Then we get an indexer of all the nodes in the active document and loop through this indexer checking whether the atoms connected by the bond are present in the particle system and adding bonds and their equilibrium length to the according vectors. The equilibrium spring length is taken as the covalent bond distances in the initial conformation of the model. Then we initialize the energy and forces to zero.
Updating the interaction model
Now, we implement the update of the interaction model.
Modify the updateInteractions function (SESpringsInteractionModel.cpp). This function is called to update the energy and forces.
Here, we reset the energy and forces, then apply the bond spring interaction model, and emit the changed signal to let SAMSON know that the energy and forces have been changed.
Implement the updateBondSpringsInteractions function as follows:
In the loop, for each spring:
- get positions of atoms connected via a bond,
- compute the force intencity as the difference of distance between the current and reference bond string (the equilibrium bond length),
- compute the force acting on atoms connected via the bond,
- sum up the energy.
You have implemented a simple interaction model. Build the Extension and launch SAMSON. Create or load any system, and apply a simulator to it (Simulation > Add or Ctrl / Cmd⌘ + Shift + M ) with the newly created interaction model and the Interactive modeling state updater:
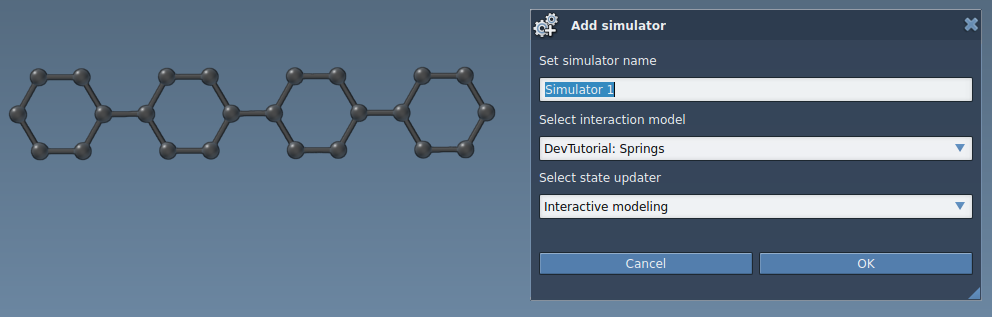
You will see property windows of the interaction model and the state updater:
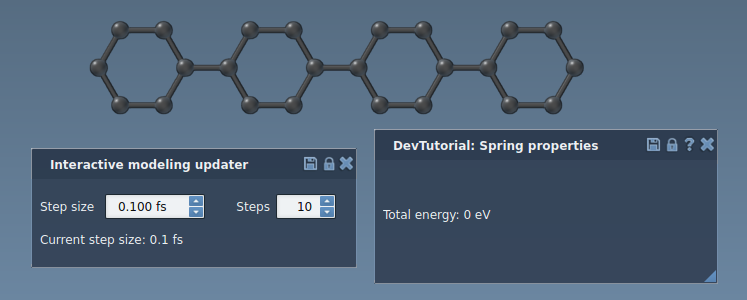
Run a simulator: press Space or the play icon in the toolbar:

The simulator should now work properly, updating the energy and forces along the simulation. Try to slightly pull an atom connected via a bond to another atom. You should see how the bond spring tries to bring another atom along with the one being pulled.
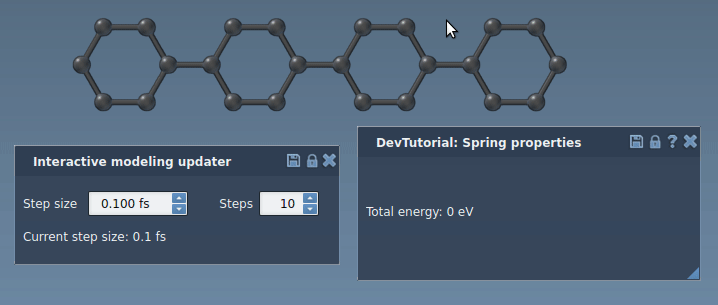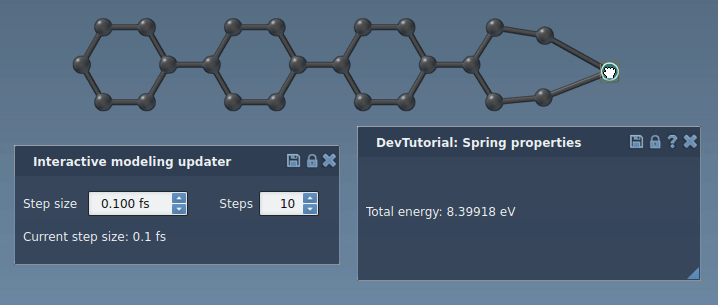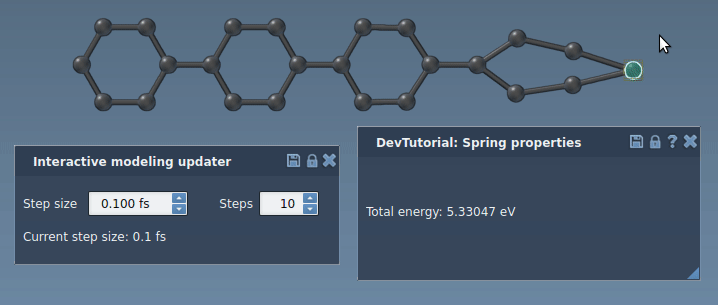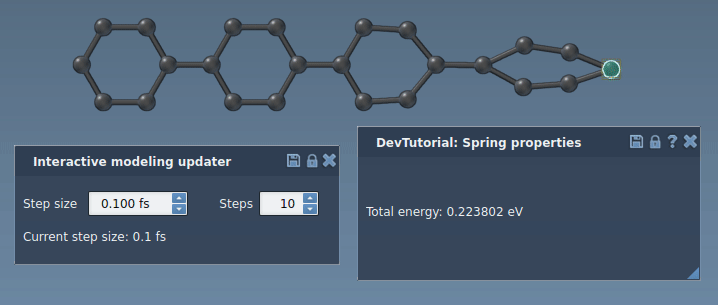
Customizing the interaction model interface
Changing the spring stiffness parameter
We want to allow the user to control the bond spring stiffness (bondSpringStiffness) through the property window of the interaction model.
Open the SESpringsInteractionModelProperties.ui, add a label and a QDoubleSpinBox, name the spin box as doubleSpinBoxSpringStiffness, set its value to 1.0, and minimum to zero, as shown on an image below:
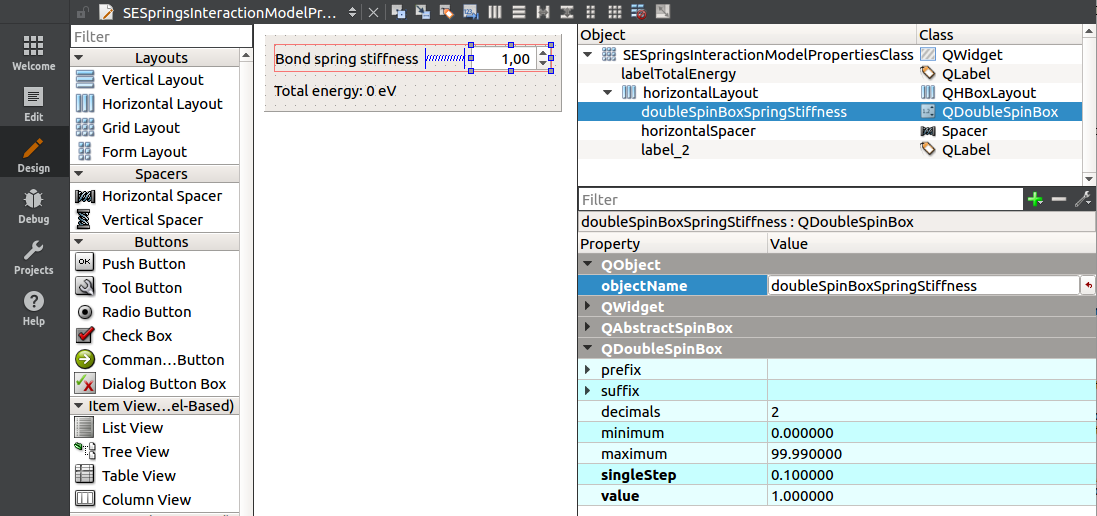
Add a slot onBondSpringStiffnessChanged(double):
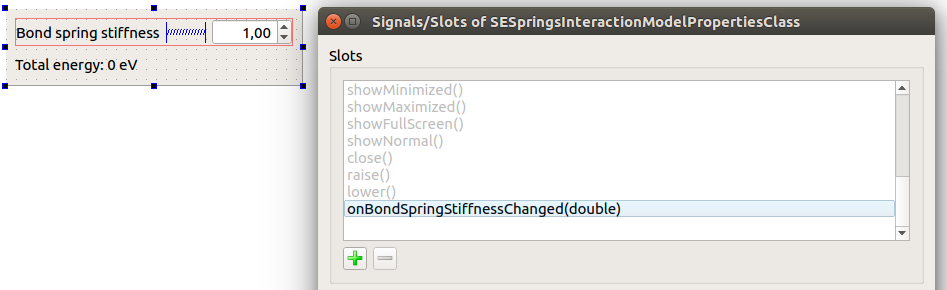
Connect the valueChanged(double) signal of the doubleSpinBoxSpringStiffness to the onBondSpringStiffnessChanged(double) slot:

Add the onBondSpringStiffnessChanged(double) slot in the SESpringsInteractionModelProperties class (SESpringsInteractionModelProperties.hpp):
and implement it as follows (SESpringsInteractionModelProperties.cpp):
Add the setBondSpringStiffness function to the SESpringsInteractionModel class (SESpringsInteractionModel.hpp):
Now, when you launch a simulator with this interaction model in SAMSON you will be able to modify the bond spring stiffness parameter through the property window. Try to pull an atom with different bond spring stiffness parameter:
If we want to save the spring stiffness parameter value for the next launch, we need to implement the saveSettings and loadSettings functions for the SESpringsInteractionModelProperties class:
Reinitializing the equilibrium bond distance
Let's now provide the user the possibility to reinitialize the springs equillibrium length.
For that, add a QPushButton to the SESpringsInteractionModelProperties.ui:
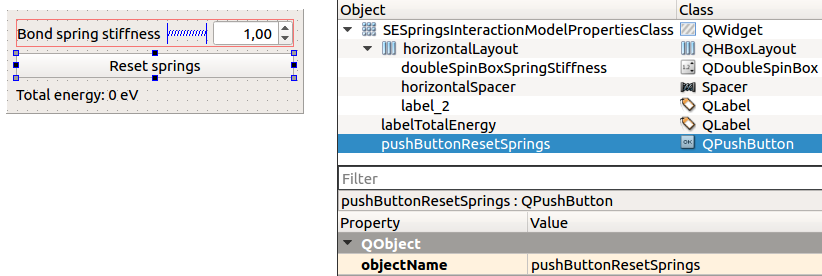
Add a slot onBondSpringStiffnessChanged(double):

Connect the valueChanged(double) signal of the doubleSpinBoxSpringStiffness to the onBondSpringStiffnessChanged(double) slot:

Add the onResetSpringsClicked slot in the SESpringsInteractionModelProperties class (SESpringsInteractionModelProperties.hpp):
and implement it as follows (SESpringsInteractionModelProperties.cpp):
Now, when you launch the simulator with this interaction model in SAMSON you will be able to reset the spring equillibrium distance through the property window. Try to pull an atom and reset the springs equillibrium distance: