Visual models are one of the five categories of models in SAMSON. They provide visual representations in the viewport. A visual model may be e.g. a secondary structure representation applicable to protein models, an electron density isosurface, a volume rendering of an electrostatics field, etc. When a visual model is loaded at startup, SAMSON stores it in a list of available visual models, used in particular when adding a visual model from the Visualization menu.
There is a number of default visual models supplied with SAMSON, you can see some of them on the image below.
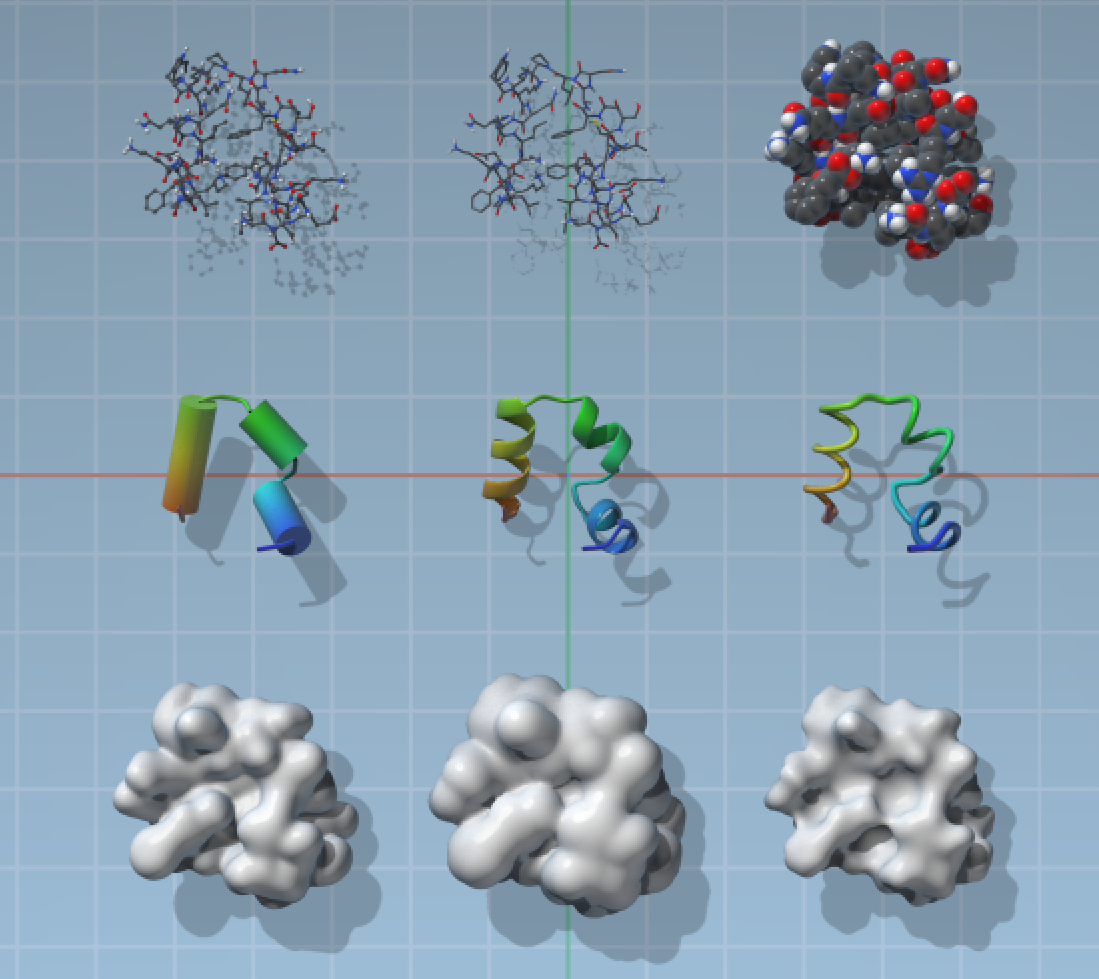
Visual models can be applied to selected nodes (or the whole document if nothing is selected) via the context menu (Add) or using Visualization menu or Biology menu.
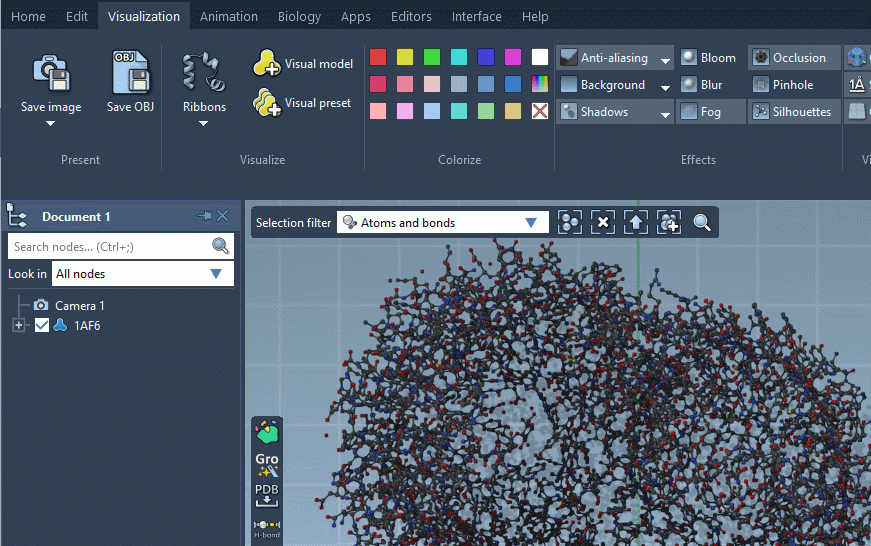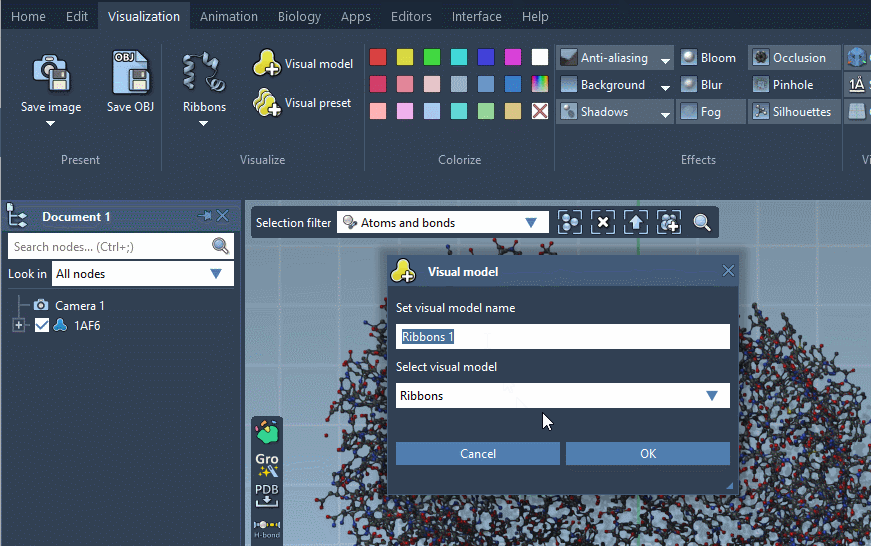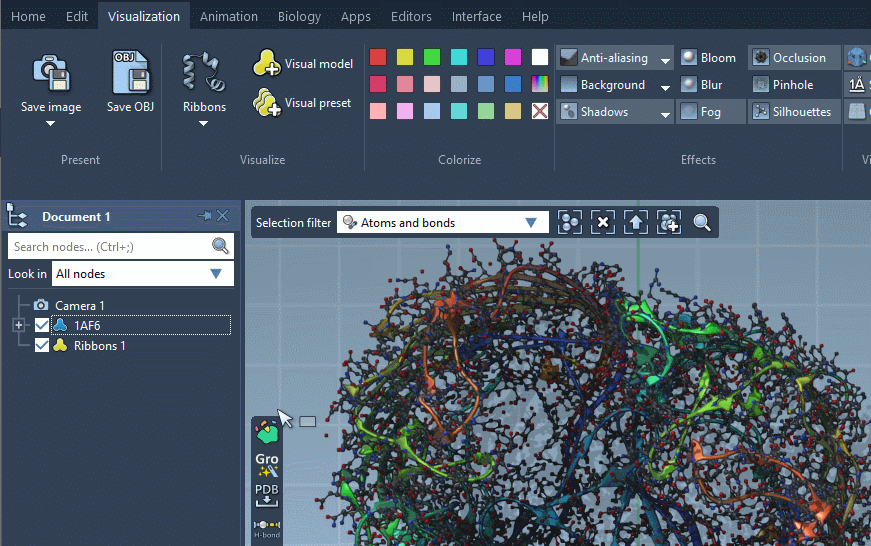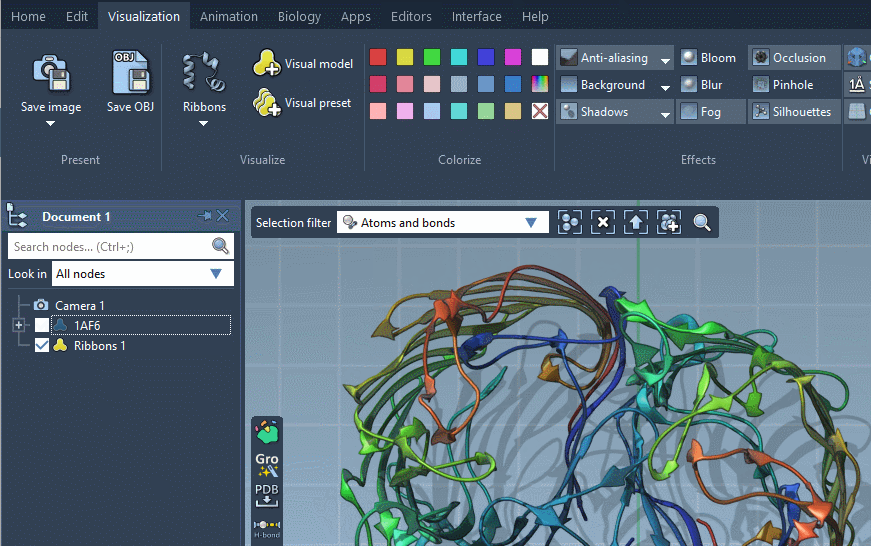
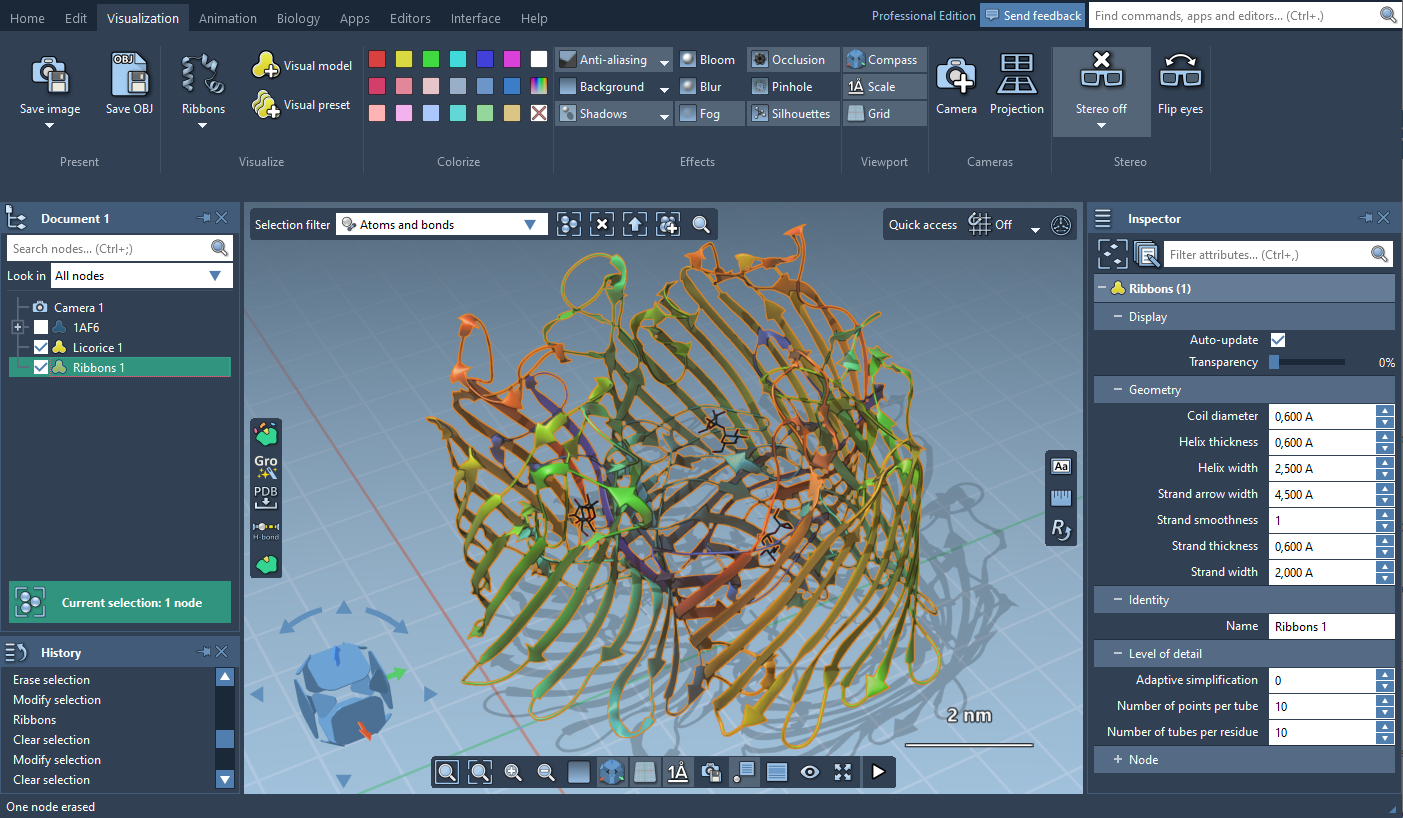
For some visual models, you can modify their visualization parameters in the Inspector. For that, select the visual model in the document view and click on Inspect in the context menu.
The default visual models in SAMSON make it possible to highlight and select atoms, residues, chains, etc. directly via surfaces.

If you want to start developing you own visual models, please, refer to the documentation about the generating SAMSON extensions and the SAMSON tutorials page for more information about writing new visual models for SAMSON.

