Using the SMILES Manager
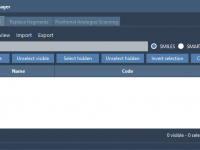
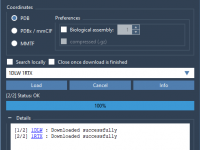
In this tutorial, you will learn how to use the SMILES Manager extension of SAMSON. This extension is based on RDKit. RDKit is a widely used open-source toolkit for cheminformatics. One of its features is the conversion of molecules SMILES strings to 2D and 3D structures. The extension interface presents three tabs: Manage SMILES, Replace fragments, and Positional Analogue Scanning. In this tutorial, we will present the first two sections one by one as they are totally independent. For the…