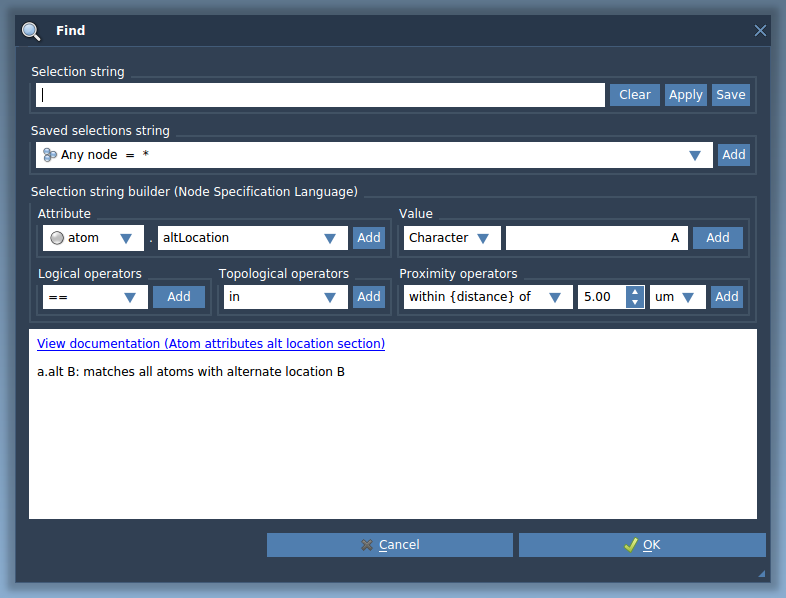
SAMSON has a powerful Node Specification Language (NSL) that may be used to select data graph nodes based on their properties. For example, the Find command of the user interface of SAMSON lets users enter a NSL string to select nodes from the document:
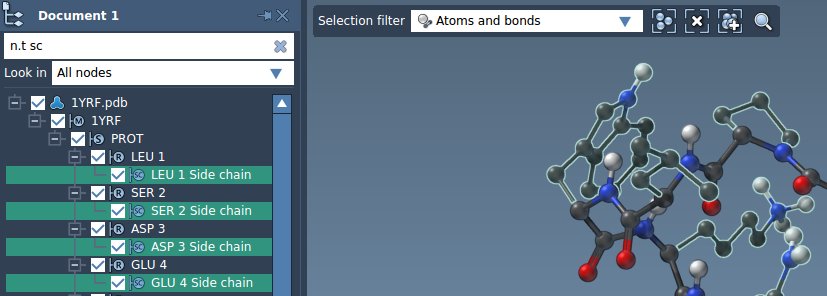
A NSL expression may also be used to filter nodes from the document view. In the example below, the user has entered n.t sc (node.type sidechain) and hit the Enter key to select all side chains:
Here are some examples of NSL expressions:
Hydrogen: select all hydrogens (short version:H)atom.chainID > 2: select all atoms with a chain ID strictly larger than 2 (short version:a.ci > 2)Carbon in node.selected: select all carbons in the current selection (short version:C in n.s)bond.order > 1.5: select all bonds with order strictly larger than 1.5 (short version:b.o > 1.5)node.type backbone: select all backbone nodes (short version:n.t bb)O in node.type sidechain: select all oxygens in side chain nodes (short version:O in n.t sc)"CA" within 5A of S: select all nodes named "CA" that are within 5 angstrom of any sulfur atom (short version:"CA" w 5A of S) (use quotes, since names may contain spaces)node.type residue beyond 5A of node.selected: select all residue nodes beyond 5 angstrom of the current selection (short version:n.t r b 5A of n.s)residue.secondaryStructure helix: select residue nodes in alpha helices (short version:r.ss h)node.type sidechain having S: select side chain nodes that have at least one sulfur atom (short version:n.t sc h S)H linking O: select all hydrogens bonded to oxygen atoms (short version:H l O)
Logical operators may be used:
C or H: select atoms that are carbons or hydrogens
When using the Find command, selections may be saved as groups in the data graph:
"Interface" = ((n.t a in "A") w 4A of "B") or ((n.t a in "B") w 4A of "A"): creates a group containing atoms at the interface between"A"and"B"
When using the Find command as well, you may press the Tab key in the search box for context-sensitive completion. This is particularly useful when searching nodes by name. For example, entering "ALA" (with the quote sign, to indicate a name) and pressing the Tab key will list all nodes whose name begins with "ALA", e.g:
"ALA 22 Backbone""ALA 22 Side-chain""ALA 22""ALA 28 Backbone""ALA 28 Side-chain""ALA 28"...
Specification by attributes
Nodes may be specified by their attributes. Since nodes may have different types in SAMSON (e.g. atom, bond, etc.), each attribute is defined in an attribute space.
The node attribute space (short name n) corresponds to attributes that are defined for each node. For example, the selection flag is a node attribute, since each node has a selection flag. Hence, the NSL expression node.selectionFlag true may match any node whose selection flag is true, regardless of its node type (atom, bond, etc.).
SAMSON's Node Specification Language supports several different attribute spaces:
node(short namen), see Node attributesatom(short namea), see Atom attributesbond(short nameb), see Bond attributeschain(short namec), see Chain attributesresidue(short namer), see Residue attributesstructuralGroup(short namesg), see Structural group attributes
Node attributes
Node attributes, defined in the node attribute space (short name n), correspond to attributes defined in each node of the data graph. For example, the selection flag is a node attribute, since each node has a selection flag. Hence, the NSL expression node.selectionFlag true may match any node whose selection flag is true, regardless of its node type (atom, bond, etc.).
Possible node attributes are:
- hasMaterial (short name
hm) - hidden (short name
h) - locked (short name
l) - lockedFlag (short name
lf) - ownsMaterial (short name
om) - selected (short name
s) - selectionFlag (short name
sf) - type (short name
t) - visibilityFlag (short name
vf) - visible (short name
v)
hasMaterial
The hasMaterial attribute (short name hm) matches nodes that have a material, either because they own it (i.e. the material is applied to them) or because they inherit it (i.e. a material is applied to one of their ascendants).
Possible values: none.
Example:
n.hm: matches all nodes which have a material
hidden
The hidden attribute (short name h) matches nodes that are hidden, either because their visibility flag is false, or because the visibility flag of one of their ancestors is false.
Possible values: none.
Example:
n.h: matches all hidden nodes
locked
The locked attribute (short name l) matches nodes that are locked, either because their locked flag is true, or because the locked flag of one of their ancestors is true.
Possible values: none.
Example:
n.l: matches all locked nodes
lockedFlag
The lockedFlag attribute (short name lf) matches nodes based on their locked flag.
Possible values: true or false.
Example:
n.lf true: matches all nodes with a locked flag set totrue
ownsMaterial
The ownsMaterial attribute (short name om) matches nodes that own a material (i.e. the material is applied to them).
Possible values: none.
Example:
n.om: matches all nodes which own a material
selected
The selected attribute (short name s) matches nodes that are selected, either because their selection flag is true, or because the selection flag of one of their ancestors is true.
Possible values: none.
Example:
n.s: matches all selected nodes
selectionFlag
The selectionFlag attribute (short name sf) matches nodes based on their selection flag.
Possible values: true or false.
Example:
n.sf true: matches all nodes with a selection flag set totrue
type
The type attribute (short name t) matches nodes by type.
Possible values:
animation(short namean)atom(short namea)backbone(short namebb)bond(short nameb)camera(short nameca)chain(short namec)conformation(short nameco)document(short named)dynamicalModelParticleSystem(short namedmps)folder(short namef)hydrogenBondGroup(short namehbg)interactionModelParticleSystem(short nameimps)label(short namela)mesh(short nameme)molecule(short namem)nodeGroup(short nameng)note(short nament)presentation(short namepr)propertyModel(short namepm)pseudoatom(short namepa)pseudobond(short namepb)residue(short namer)segment(short names)sidechain(short namesc)simulatorParticleSystem(short namesps)stateUpdaterParticleSystem(short namesups)structuralGroup(short namesg)structuralModel(short namesm)structuralParticle(short namesp)structuralRoot(short namesr)visualModel(short namevm)
Examples:
n.t a: matches all atomsn.t sc: matches all side-chain nodesn.t vm: matches all visual models
visibilityFlag
The visibilityFlag attribute (short name vf) matches nodes based on their visibility flag.
Possible values: true or false.
Example:
n.vf true: matches all nodes with a visibility flag set totrue
visible
The visible attribute (short name v) matches nodes that are visible, i.e. the nodes whose visibility flag is true and whose ancestors are visible.
Possible values: none.
Example:
n.v: selects all visible nodes
Atom attributes
Atom attributes are defined in the atom attribute space (short name a). Atom attributes may only match atom nodes.
Possible atom attributes are:
- altLocation (short name
alt) - aminoAcidBackbone (short name
aabb) - aromatic (short name
ar) - chain (short name
c) - chainID (short name
ci) - customType (short name
ct) - element (short name
e) - formalCharge (short name
fc) - geometry (short name
g) - hetatm (short name
het) - hybridization (short name
hy) - metal (short name
met) - mobile (short name
mo) - nucleicAcidBackbone (short name
nabb) - numberOfBondedAtoms (short name
nba) - numberOfBondedCarbons (short name
nbc) - numberOfBondedHeavyAtoms (short name
nbha) - numberOfBondedHydrogens (short name
nbh) - numberOfBondedNitrogens (short name
nbn) - numberOfBondedOxygens (short name
nbo) - numberOfBondedSulfurs (short name
nbs) - occupancy (short name
oc) - partialCharge (short name
pc) - planar (short name
pl) - residueSequenceNumber (short name
resi) - resonance (short name
reso) - serialNumber (short name
sn) - sybyl (short name
sy) - symbol (short name
s) - temperatureFactor (short name
tf) - water (short name
w) - x
- y
- z
altLocation
The altLocation attribute (short name alt) matches atoms based on their alternate location.
Possible values: a character.
Example:
a.alt B: matches all atoms with alternate location B
aminoAcidBackbone
The aminoAcidBackbone attribute (short name aabb) matches atoms that belong to an amino-acid backbone.
Possible values: none.
Example:
a.aabb: matches atoms that belong to an amino-acid backbone
aromatic
The aromatic attribute (short name ar) matches atoms that are aromatic.
Possible values: none.
Example:
a.ar: matches aromatic atoms
chain
The chain attribute (short name c) matches atoms that are aromatic.
Possible values: a character.
Example:
a.c A: matches atoms from chain A
chainID
The chainID attribute (short name ci) matches atoms with specific chain IDs.
Possible values: an integer.
Example:
a.ci == 0: matches atoms from chain ID 0a.ci >= 0: matches atoms from chain ID larger than 0a.ci >= 0 and a.ci <=2: matches atoms from chain ID between 0 and 2
customType
The customType attribute (short name ct) matches atoms with specific custom types.
Possible values: an integer.
Example:
a.ct == 0: matches atoms with custom type 0a.ct >= 0: matches atoms with custom type larger than 0a.ct >= 0 and a.ct <=2: matches atoms with custom type between 0 and 2
element
The element attribute (short name e) matches atoms by element.
Possible values: element names.
Example:
a.e Carbon: matches carbon atomsa.e Nitrogen: matches nitrogen atoms
formalCharge
The formalCharge attribute (short name fc) matches atoms with specific formal charges.
Possible values: integer values.
Example:
a.fc >= 1: matches atoms with formal charge larger than 1
geometry
The geometry attribute (short name g) matches atoms with specific geometry. Please note, that geometry needs to be computed first.
Possible geometry type values:
linearbenttrigonalplanartrigonalpyramidaltshapedtetrahedralsquareplanarseesawtrigonalbipyramidalsquarepyramidalpentagonalplanaroctahedraltrigonalprismaticpentagonalpyramidalpentagonalbipyramidalcappedoctahedralcappedtrigonalprismaticsquareantiprismaticdodecahedralbicappedtrigonalprismatictricappedtrigonalprismaticcappedsquareantiprismaticundefined
Example:
a.g tet: matches atoms with tetrahedral geometry
hetatm
The hetatm attribute (short name het) matches heteroatoms in protein structures, i.e. atoms whose record type is HETATM in the Protein Data Bank file format.
Possible values: none.
Example:
a.het: matches heteroatoms
hybridization
The hybridization attribute (short name hy) matches atoms with specific hybridization. Please note, that hybridization needs to be assigned first.
Possible hybridization type values:
noneSPSP2SP3SP3DSP3D2unknown
Example:
a.hy SP2: matches atoms with SP2 hybridization.
metal
The metal attribute (short name met) matches atoms from metal subcategories.
Possible values: none.
Example:
a.met: matches atoms from metal subcategories
mobile
The mobile attribute (short name mo) matches atoms that are mobile.
Possible values: none.
Example:
a.mo: matches mobile atoms
nucleicAcidBackbone
The nucleicAcidBackbone attribute (short name nabb) matches atoms that belong to a nucleic acid backbone.
Possible values: none.
Example:
a.nabb: matches atoms that belong to a nucleic acid backbone
numberOfBondedAtoms
The numberOfBondedAtoms attribute (short name nba) matches atoms with a specific number of bonded atoms.
Possible values: integers.
Example:
a.nba > 3: matches atoms that have more than 3 bonded atoms
numberOfBondedHeavyAtoms
The numberOfBondedHeavyAtoms attribute (short name nbha) matches atoms with a specific number of bonded heavy (non-hydrogen) atoms.
Possible values: integers.
Example:
a.nbha == 3: matches atoms that have exactly 3 bonded heavy atoms
numberOfBondedCarbons
The numberOfBondedCarbons attribute (short name nbc) matches atoms with a specific number of bonded carbon atoms.
Possible values: integers.
Example:
a.nbc > 3: matches atoms that have more than 3 bonded carbon atoms
numberOfBondedHydrogens
The numberOfBondedHydrogens attribute (short name nbh) matches atoms with a specific number of bonded hydrogen atoms.
Possible values: integers.
Example:
a.nbh == 3: matches atoms that have exactly 1 bonded hydrogen atom
numberOfBondedNitrogens
The numberOfBondedNitrogens attribute (short name nbn) matches atoms with a specific number of bonded nitrogen atoms.
Possible values: integers.
Example:
a.nbn < 2: matches atoms that have less than 2 bonded nitrogen atoms
numberOfBondedOxygens
The numberOfBondedOxygens attribute (short name nbo) matches atoms with a specific number of bonded oxygen atoms.
Possible values: integers.
Example:
a.nbo == 2: matches atoms that have exactly 2 bonded oxygen atoms
numberOfBondedSulfurs
The numberOfBondedSulfurs attribute (short name nbs) matches atoms with a specific number of bonded sulfur atoms.
Possible values: integers.
Example:
a.nbs == 0: matches atoms that have zero bonded sulfur atoms
occupancy
The occupancy attribute (short name oc) matches atoms with a specific occupancy.
Possible values: floating-point values.
Example:
a.oc >= 2: matches atoms with occupancy larger than 2
partialCharge
The partialCharge attribute (short name pc) matches atoms with specific partial charges.
Possible values: floating-point values.
Example:
a.pc >= 1.3: matches atoms with partial charge larger than 1.3
planar
The planar attribute (short name pl) matches planar atoms, i.e. atoms that are in a plane with its covalently bonded atoms.
Possible values: none.
Example:
a.pl: matches planar atoms
residueSequenceNumber
The residueSequenceNumber attribute (short name resi) matches atoms in residues with specific indices.
Possible values: integers.
Example:
a.resi == 12: matches atoms in residue 12a.resi >= 12 and a.resi <=97: matches atoms in residue 12 to 97
resonance
The resonance attribute (short name reso) matches resonant atoms.
Possible values: none.
Example:
a.reso: matches resonant atoms
serialNumber
The serialNumber attribute (short name sn) matches atoms with specific serial numbers.
Possible values: integers.
Example:
a.sn >= 20: matches atoms with serial number larger than 20a.sn >= 20 and a.sn<=897: matches atoms with serial number between 20 and 897
sybyl
The sybyl attribute (short name sy) matches atoms with the specified SYBYL type. Please note, that atoms need to have SYBYL types assigned.
Possible values: sybyl type names, e.g. C.2, C.3, N.2, etc.
Example:
a.sy C.3: matches atoms with the specified SYBYL type
symbol
The symbol attribute (short name s) matches atoms with specific symbols.
Possible values: element symbols.
Example:
a.s C: matches carbon atomsa.s C or a.s H: matches atoms that are carbons or hydrogens
temperatureFactor
The temperatureFactor attribute (short name tf) matches atoms with specific temperature factors.
Possible values: floating-point values.
Example:
a.tf > 2: matches atoms with a temperature factor strictly larger than 2
water
The water attribute (short name w) matches water atoms.
Possible values: none.
Example:
a.w: matches water atoms
x
The x attribute matches atoms with specific x coordinates
Possible values: floating-point values with length units.
Example:
a.x >= 1.0 A: matches atoms whose x coordinate is larger than 1.0 A
Possible units for lengths are:
micrometer(short nameum)nanometer(short namenm)angstrom(short nameA)picometer(short namepm)femtometer(short namefm)
y
The y attribute matches atoms with specific y coordinates
Possible values: floating-point values with length units.
Example:
a.y >= 1.0 A: matches atoms whose y coordinate is larger than 1.0 A
Possible units for lengths are:
micrometer(short nameum)nanometer(short namenm)angstrom(short nameA)picometer(short namepm)femtometer(short namefm)
z
The z attribute matches atoms with specific z coordinates
Possible values: floating-point values with length units.
Example:
a.z >= 1.0 A: matches atoms whose z coordinate is larger than 1.0 A
Possible units for lengths are:
micrometer(short nameum)nanometer(short namenm)angstrom(short nameA)picometer(short namepm)femtometer(short namefm)
Bond attributes
Bond attributes are defined in the bond attribute space (short name b). Bond attributes may only match bond nodes.
Possible bond attributes are:
- customType (short name
ct) - length (short name
len) - order (short name
o)
customType
The customType attribute (short name ct) matches bonds with specific custom types.
Possible values: an integer.
Example:
b.ct == 0: matches bonds with custom type 0b.ct >= 0: matches bonds with custom type larger than 0b.ct >= 0 and b.ct <=2: matches bonds with custom type between 0 and 2
length
The length attribute (short name len) matches bonds with specific bond length.
Possible values: floating-point values.
Example:
b.len >= 1.5A: matches all bonds with length larger than 1.5 angstroms
order
The order attribute (short name o) matches bonds with specific orders.
Possible values: floating-point values.
Example:
b.o >= 2: matches all bonds with order larger than 2
Chain attributes
Chain attributes are defined in the chain attribute space (short name c). Chain attributes may only match chain nodes.
Possible chain attributes are:
- chainID (short name
id)
chainID
The chainID attribute (short name id) matches chains with specific chain ID.
Possible values: integers.
Example:
c.id == 1: matches all chains with chain ID equal to 1
Residue attributes
Residue attributes are defined in the residue attribute space (short name r). Residue attributes may only match residue nodes.
Possible residue attributes are:
- aminoAcid (short name
aa) - charge (short name
c) - completeAminoAcidBackbone (short name
caab) - nucleicAcid (short name
na) pKa1,pKa2, andisoelectricPointPH(short namepI), see Dissociation constants- polarity (short name
p) - residueSequenceNumber (short name
id) - secondaryStructure (short name
ss) - terminal (short name
ter) - type (short name
t)
aminoAcid
The aminoAcid attribute (short name aa) matches residues that are amino acids.
Possible values: boolean.
Example:
r.aa: matches all residues that are amino acids
charge
The charge attribute (short name c) matches amino acid residues with specific charge.
Possible values:
negativeneutralpositive
Example:
r.c negative: matches all amino acid residues with negative side chain charge
completeAminoAcidBackbone
The completeAminoAcidBackbone attribute (short name caab) matches residues that have complete amino acid backbones.
Possible values: boolean.
Example:
r.hcaab: matches all residues that have complete amino acid backbones
nucleicAcid
The nucleicAcid attribute (short name na) matches residues that are nucleic acids.
Possible values: boolean.
Example:
r.na: matches all residues that are nucleic acids
Dissociation constants
The pKa1, pKa2, and isoelectricPointPH (short name pI) attributes matches amino acid residues with certain dissociation constants: pKa1 - the negative of the logarithm of the dissociation constant for the carboxyl functional group, -COOH pKa2 - the negative of the logarithm of the dissociation constant for the amino functional group, -NH3 pI - the pH at the isoelectric point
Reference: D.R. Lide, Handbook of Chemistry and Physics, 72nd Edition, CRC Press, Boca Raton, FL, 1991.
Possible values: floating-point values.
Example:
r.pKa1 < 2.0: matches amino acid residues with pKa1 values less than 2
polarity
The polarity attribute (short name p) matches amino acid residues with specific polarity.
Possible values:
acidicPolar(alsoacidic)basicPolar(alsobasic)nonpolarpolar
Example:
r.p polar: matches all amino acid residues with a polar side chain
residueSequenceNumber
The residueSequenceNumber attribute (short name id) matches residues with specific residue sequence number (structure ID).
Possible values: integers.
Example:
r.id > 1 and r.id < 10: matches all residues with residue sequence number between 1 and 10
secondaryStructure
The secondaryStructure attribute (short name ss) matches residues with specific secondary structures.
Possible values:
alpha(short namea)beta(short nameb)unstructured(short nameu)helix(short nameh) (same matches asalpha)strand(short names) (same matches asbeta)loop(short namel) (same matches asunstructured)
Example:
r.ss h: matches all residues in alpha helices
terminal
The terminal attribute (short name ter) matches residues that are terminal.
Possible values: boolean.
Example:
r.ter: matches all residues that are terminal
type
The type attribute (short name t) matches residues with specific types.
Possible values:
A,C,G,U,IDA,DC,DG,DT,DIALA,ARG,ASP,ASN,VAL,HIS,GLY,GLU,GLN,ILE,LEU,LYS,MET,PRO,SER,TYR,THR,TRP,PHE,CYS,ASX,GLX,XLE,XAA,SEC,PYL
Example:
r.t ALA: matches all alanines
Structural group attributes
Structural group attributes are defined in the structuralGroup attribute space (short name sg). Structural group attributes may only match structural group nodes.
Possible structural group attributes are:
- structureID (short name
id)
structureID
The structureID attribute (short name id) matches structural groups with specific structure ID.
Possible values: integers.
Example:
sg.id == 1: matches all structural groups with structure ID equal to 1
Specification by name
Nodes may be specified by names, i.e. strings enclosed with quotes. In the SAMSON data graph, nodes which may have a custom name are:
- animations
- atoms
- backbones
- cameras
- chains
- conformations
- dynamical models
- hydrogen bond groups
- groups
- interaction models
- labels
- folders
- molecules
- presentations
- property models
- residues
- segments
- side chains
- simulators
- structural groups
- structural models
- visual models
- meshes
Note that bond names are formed from the names of the atoms they bond, and cannot be searched by name.
As noted above, pressing the Tab key in the search box when a string is being entered (with the quote sign, to indicate a name) lists all nodes whose name begins with the entered string. For example, entering "ALA" and pressing the Tab key may yield e.g:
"ALA 22 Backbone""ALA 22 Side-chain""ALA 22""ALA 28 Backbone""ALA 28 Side-chain""ALA 28"...
Possible values: a string enclosed with quotes.
Examples:
"Folder 1""CA""Group 1""1EQZ""ALA 28""Nanotube""Simulator 1"
Specification by symbols and element names
In order to efficiently match atoms of a given element type, symbols and element names are valid NSL expressions.
Possible values: atomic symbols and element names (capitalized)
Examples:
Carbon: matches all carbon atomsC: matches all carbon atomsCa: matches all calcium atomsH: matches all hydrogen atoms
Operations on sets
Logical operators
Logical operators may be used to perform operations on sets.
Possible values: and, not, or and xor (exclusive or).
Examples:
a.sn >= 20 and a.sn <= 897: matches atoms with serial number between 20 and 897a.sn <= 20 or a.sn >=40: matches atoms with serial number smaller than 20 or larger than 40n.t r and not r.t CYS: matches residue nodes that are not cysteinsa.sn >= 20 xor a.oc >= 0.5: matches atoms which either have a serial number larger than 20, or have an occupancy larger than 0.5, but not those that satisfy both conditions
Note that, without proper care, the not condition might produce surprising results. For example, not r.t CYS does not return the list of residues that are not cysteins. Indeed, a folder node is also a node which is not a residue node that's a cystein. If only residues nodes should be returned, a proper query would be n.t r and not r.t CYS.
Topology operators
Containement operators may be used to specify inclusion in or out of a set.
Possible values: in and out of.
Examples:
n.t a in "2AZ8": matches atoms in "2AZ8"n.t a in r.t CYS: matches atoms that belong to cysteinsH in r.t ARG: matches hydrogens that belong to argininsH in n.h: matches hidden hydrogensn.t a out of r.t PRO: matches atoms that do not belong to prolines
Proximity operators
Proximity operators may be used to select nodes based on distances.
Possible values: within {distance} of and beyond {distance} of.
Examples:
C within 5A of "GLN 2": matches carbons within 5 angstrom of "GLN 2"n.t a beyond 5A of "2AZ8-IA": matches atoms beyond 5 angstrom of "2AZ8-IA"
Possible units for distances are:
micrometer(short nameum)nanometer(short namenm)angstrom(short nameA)picometer(short namepm)femtometer(short namefm)
Note that, without proper care, the within condition might produce surprising results. For example, * within 5A of "GLN 2" will select the document node, since some atoms in the document are within 5 angstrom of "GLN 2": the atoms that belong to "GLN 2". If only atom nodes should be returned, a proper query would be n.t a within 5A of "GLN 2". Note that this latter query would also return atoms that belong to "GLN 2". If only atoms outside "GLN 2" should be returned, then a proper query would be n.t a within 5A of "GLN 2" out of "GLN 2".
Managing groups
As noted above, selections may be saved as groups in the data graph. For example, "Interface" = ((n.t a in "A") w 4A of "B") or ((n.t a in "B") w 4A of "A") creates a group containing atoms at the interface between "A" and "B". Precisely, a group node called "Interface" is added to the document.
Note that "node group" is a specific node type. As a result, the query "Interface" will return the group node itself, and not the nodes forming the group.
Reference
Attributes
Node attributes
| Attribute name | Short name | Possible values | Examples |
hasMaterial | hm | n.hm | |
hidden | h | n.h | |
ownsMaterial | om | n.om | |
selected | s | n.s | |
selectionFlag | sf | true or false | n.sf true |
type | t | See node type | n.t a |
visibilityFlag | vf | true or false | n.vf false |
visible | v | n.v |

