Modeling and simulation are two of the main tasks performed in SAMSON.
Models and simulators are nodes in SAMSON Document.
Models
SAMSON represents nanosystems using five categories of models:
- Structural models describe geometry and topology
- Visual models provide graphical representations
- Dynamical models describe dynamical degrees of freedom
- Interaction models describe energies and forces
- Property models describe properties that do not enter in the first four model categories
Structural models
Structural models describe the geometry and topology of nanosystems in SAMSON. Typically, a structural model contains atoms and bonds, and might contain nodes representing higher organization levels (e.g. molecules, residues, etc.).
Visual models
Visual models are used to provide graphical representations. A visual model may be e.g. a secondary structure representation of a protein, an isosurface of an electron density, a volumetric representation of an electrostatic field, etc.
SAMSON Extensions may provide visual models that may be added to the document from the Visualization menu or via shortcut Ctrl/ Cmd⌘ + Shift + V (see SAMSON interface and the Visualizing section).
Dynamical models
Dynamical models are used to indicate where degrees of freedom are in structural nodes. For example, a particle system dynamical model applied to a group of structural particles (atoms or pseudo-atoms) assigns three translational degrees of freedom to each particle. Dynamical models describe the dynamical state and store positions, momenta and masses.
Interaction models
Interaction models are used to represent energies and forces in a dynamical model. For example, an interaction model may be e.g. a simple Lennard-Jones model to describe unbounded interactions between a group of particles, a spring model, an elastic network model, a universal force field, etc.
An interaction model is in charge of providing forces corresponding to the degrees of freedom in a dynamical model. For example, an interaction model applied to a particle system dynamical model provides both the total energy of the particle system and the force applied to each particle in the particle system.
Property models
Property models are used to represent properties of nanosystems that are not already described by the first four categories of models. For example, property models may be e.g. a simple number describing the gyration radius of a protein, a scalar field (such as an electron density), a vector field (to represent e.g. an electrostatic field), a function, etc.
Simulators
Simulators (potentially interactive ones) are used to build physically-based models, and predict properties.
A simulator is applied to a dynamical model, an interaction model, and a State updater. When users start interactive simulation, SAMSON goes through all simulators added to the data graph and calls their state updaters to update the state of the attached dynamical models. The simulator contains dynamical model, interaction model, State updater, and synchronizes these different models.
In SAMSON, force fields are named Interaction model and integrators are named state updaters. Simulations in SAMSON are interactive in the sense that users can still act on the atoms while the simulation is running.
Applying simulators
Let's try to apply a Simulators simulator to a simple system, to see how the simulation affects its geometry. Please, open the Methane molecule provided in the Samples / Hydrocarbons folder.
To add a simulator go to the Home menu and select Add Simulator or use the shortcut Ctrl/ Cmd⌘ + Shift + M. Select ASED-MO as the Interaction model and Interactive modeling as the State updater, then press Ok:
Note: The ASED-MO force field [doi:10.1002/qua.560490504, doi:10.1002/jcc.22905] is based on the semi-empirical electron-delocalization molecular-orbital theory and has been applied to different fields, including surface science and electrochemistry.
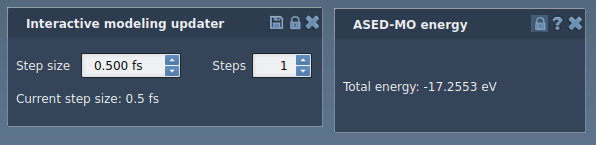
You should see two property windows: one for the Interaction model and another one for the State updater:
To launch the simulation, either go to the Home menu and click on the Start simulation or push the start simulation icon or press the Space button. To stop the simulation, either click on the Stop simulation in the Home menu or push the stop simulation icon or press the Space button.
The newly created Simulators simulator including the interaction model, State updater, and the associated dynamical model will be added in the document view:
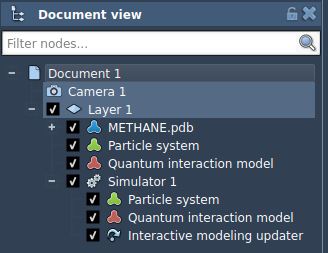
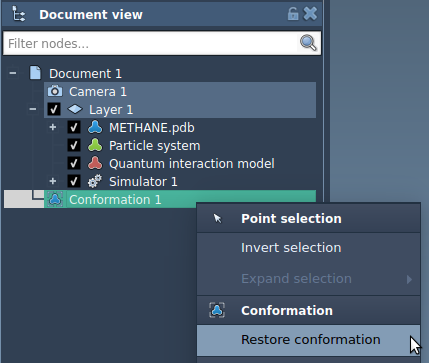
In the image above you can see the following:
- METHANE.pdb - a structural model.
- Particle system - a dynamical model to which the simulator is applied. It contains the degrees of freedom of your system.
- Quantum interaction model - an interaction model (force field) used in the simulator. It contains the computed energy and forces.
- Simulator with references to the dynamical model, interaction model, and the Interactive modeling State updater.
Select an atom in the Methane molecule and try to drag it slowly, you should see the interactive simulation in action - the molecule should follow the dragged atom.
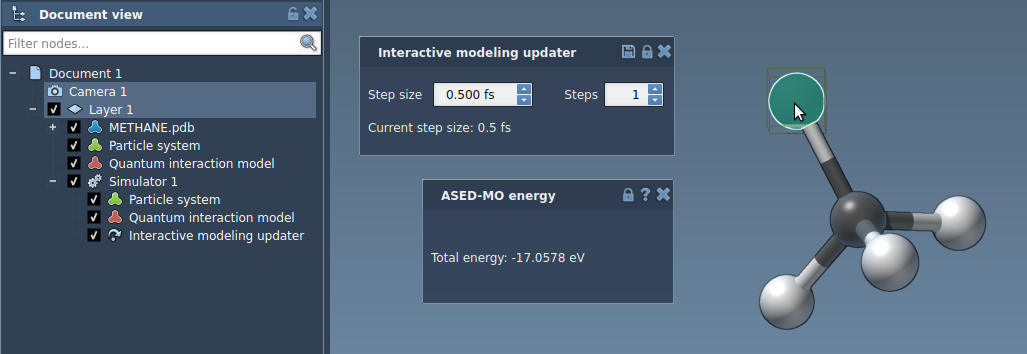
For the Interactive modeling State updater you can modify the step size and the number of steps. You can increase the number of simulations steps to increase the stiffness of the system. Try modifying the number of steps and the step size for the Interactive modeling State updater.
Saving conformations
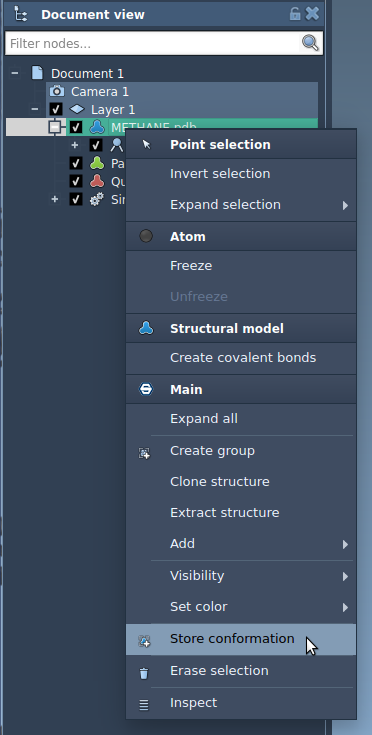
To save the current state of a structural model or group of atoms, select them and right-click on the selection, in the context menu choose Store conformation:
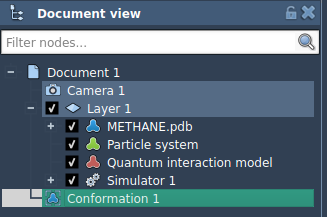
A new conformation node should appear in the document view:
You can restore a conformation by double-clicking on it in the document view or right-click and choose Restore conformation:
You can save all the conformations together with the associated structural models in a .sam or .samx file and load them in SAMSON later.

