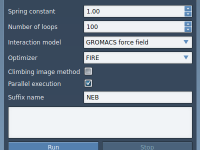
Interactive Ramachandran Plot
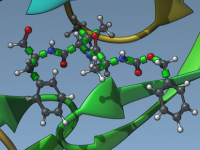
In this tutorial, we will show you how to use the Interactive Ramachandran Plot SAMSON extension. First, go to SAMSON Extensions web page, log in, and add the Interactive Ramachandran Plot SAMSON extension. The Ramachandran plot is a way to visualize energetically allowed regions for backbone dihedral angles ψ against φ of amino acid residues in protein structure. For this tutorial, you can open any protein you like. We will be using 1YRF. You can use the Protein Data Bank Downloader to fetch molecules from…