Creation of a carbon nanotube fabric: replication, modification of positions#

In this tutorial, we will weave a carbon nanotube (CNT) fabric, numerically, of course. To do so, we need to perform the following steps:
create one nanotube using the Nanotube Creator Editor;
replicate this nanotube in two directions to create a grid of nanotubes;
apply sine function to these nanotubes in the third direction to weave them with each other.
To create a nanotube, we will use the Nanotube Creator Editor.
For the sake of simplicity, we will create a nanotube in the X-direction with the size of 400 Å. To create a nanotube with specific parameters, double-click on the Nanotube Creator Editor’s icon and set parameters as on the image below.
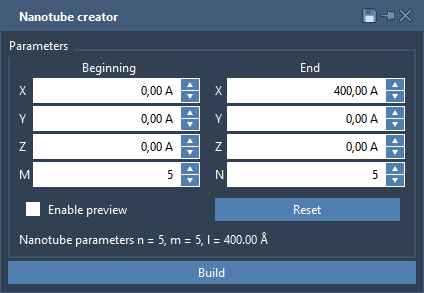
To generate a nanotube with the given parameters, press the Build button.
The rest will be done using Python. Now, let’s open the Python Scripting app.
The nanotube created with the Nanotube Creator Editor is stored in a structural model in the data graph. For the simplicity, let’s assume that we have only one nanotube created in the active document. Then, to get this nanotube object in Python, we need to get the first structural model in the active document:
# get an indexer of all structural models in the active document
indexerOfStructuralModels = SAMSON.getNodes('n.t sm')
# get the first structural model from the indexer
nanotube = indexerOfStructuralModels[0]
Let us now create a nanotube fabric by replicating the created nanotube in XY-plane and modifying Z-coordinate of atoms in nanotubes to weave these nanotubes with each other.
Before setting up parameters for the fabric, we need to find a bounding box of the created nanotube:
# get an indexer of all atoms in the nanotube
indexerOfAtoms = nanotube.getNodes('n.t a')
a = indexerOfAtoms[0]
minX = maxX = a.getX()
minY = maxY = a.getY()
# go through all atoms in the indexer
for a in indexerOfAtoms:
x = a.getX()
y = a.getY()
minX = min(minX, x)
maxX = max(maxX, x)
minY = min(minY, y)
maxY = max(maxY, y)
Let’s now set up parameters for the creation of nanotube fabric. We will set the number of replicas in one direction (the number of replicas in the perpendicular direction will be the same), and based on it we will compute the distance between replicas. You can do it differently by specifying the distance between nanotubes and then computing the number of replicas. We will be using the sine function for modifying nanotubes with its half-period set equal to the distance between nanotubes and an amplitude equal to 1.5 of the nanotube’s radius.
length = (maxX - minX) # the nanotube's length
diameter = (maxY - minY) # the nanotube's diameter
numReplicasX = 10 # the number of replicas in one direction
distance = length / numReplicasX # set еру distance between nanotubes based on the number of replicas
A = 0.75 * diameter # set an amplitude for sine function
Now, let’s replicate the nanotube in the Y-direction by copying it and shifting the copies in the Y-direction. The replicas will be added to the active document.
# get the active document
document = SAMSON.getActiveDocument()
# start holding to allow for undo/redo
with SAMSON.holding("Replicate nanotubes"):
for r in range(numReplicasX-1):
# create a replica
replica = nanotube.clone() # clone the original nanotube
SAMSON.hold(replica) # hold the replica node for undo/redo
replica.create() # create the replica
document.addChild(replica) # add the replica to the document
# shift the replica in Y-direction
dy = (r + 1) * distance
# get an indexer of all atoms in the replicated nanotube
indexerOfAtoms = replica.getNodes('n.t a')
# loop over all atoms in the replicated nanotube
for a in indexerOfAtoms:
# shift atoms in the Y-direction
a.setY(a.getY() + dy)
After doing that, we will get a set of nanotubes looking like this:
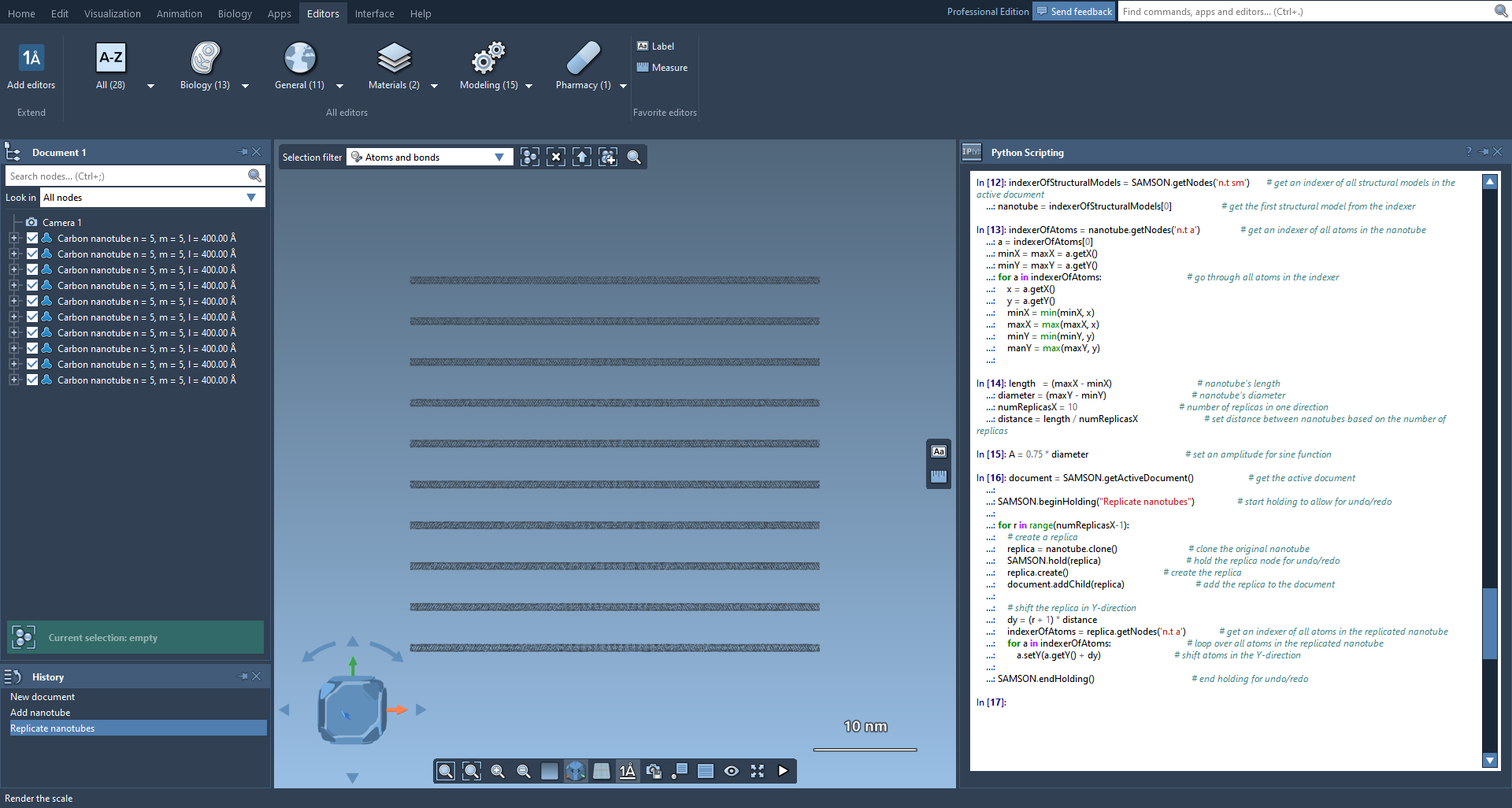
Now, we will replicate these nanotubes in the X-direction by copying them and rotating the replicas by 90 degrees. These replicas will also be shifted from the boundaries to have a grid pattern. Then, we will apply the sine function in Z-direction to all nanotubes with changing its phase to have a weaved pattern.
# import the necessary functions and constants
from math import sin, pi
# get an indexer of all structural models in the active document
indexerOfStructuralModels = SAMSON.getNodes('n.t sm')
sign = 1 # used to change the phase of the sine function
# start holding to allow for undo/redo
SAMSON.beginHolding("Replicate nanotubes")
# loop through all structural models (all nanotubes)
for original in indexerOfStructuralModels:
# create a replica
replica = original.clone() # clone the original nanotube
SAMSON.hold(replica) # hold the replica node for undo/redo
replica.create() # create the replica
document.addChild(replica) # add it to the document
# for shifting nanotubes from the border
shift = distance / 2.0
# get an indexer of all atoms in the replicated nanotube
indexerOfAtomsInReplica = replica.getNodes('n.t a')
# rotate the replica by 90 degrees by changing x and y coordinates of its atoms
# and shift the replica from the borders to have '#' pattern
for a in indexerOfAtomsInReplica:
x = a.getX()
y = a.getY()
a.setY(x - shift)
a.setX(y + shift)
# apply sine function in Z-direction to original and replicated nanotubes
# get an indexer of all atoms in the original nanotube
indexerOfAtomsInOriginal = original.getNodes('n.t a')
# loop over all atoms in the original nanotube
for a in indexerOfAtomsInOriginal:
w = a.getX() / distance # [dimensionless quantity]
# apply sine function to the Z-coordinate of an atom
a.setZ(a.getZ() + A * sign * sin(pi * w.value))
# loop over all atoms in the replicated nanotube
for a in indexerOfAtomsInReplica:
# [dimensionless quantity], take into account the shift we did previously
w = (a.getY() - shift) / distance
# apply sine function to the Z-coordinate of an atom
a.setZ(a.getZ() + A * sign * sin(pi * w.value))
# change the phase to the opposite one
sign = -1 * sign
# end holding for undo/redo
SAMSON.endHolding()
After that, we will get a grid of nanotubes, where nanotubes are woven with each other.
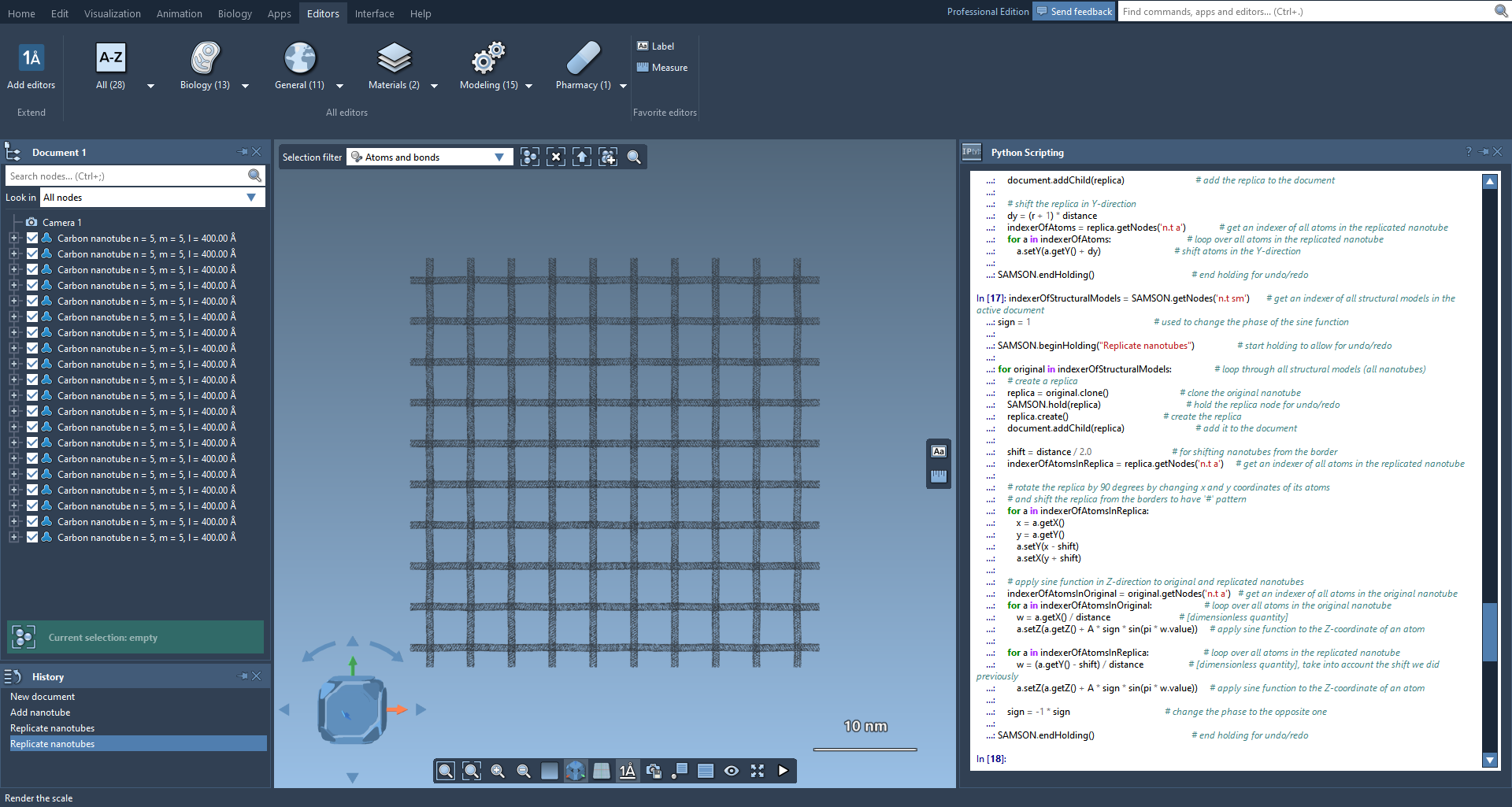
A close-up of the created system. For this picture, the default bond radius was set equal to the default atom radius in Preferences → Rendering → Structural model.
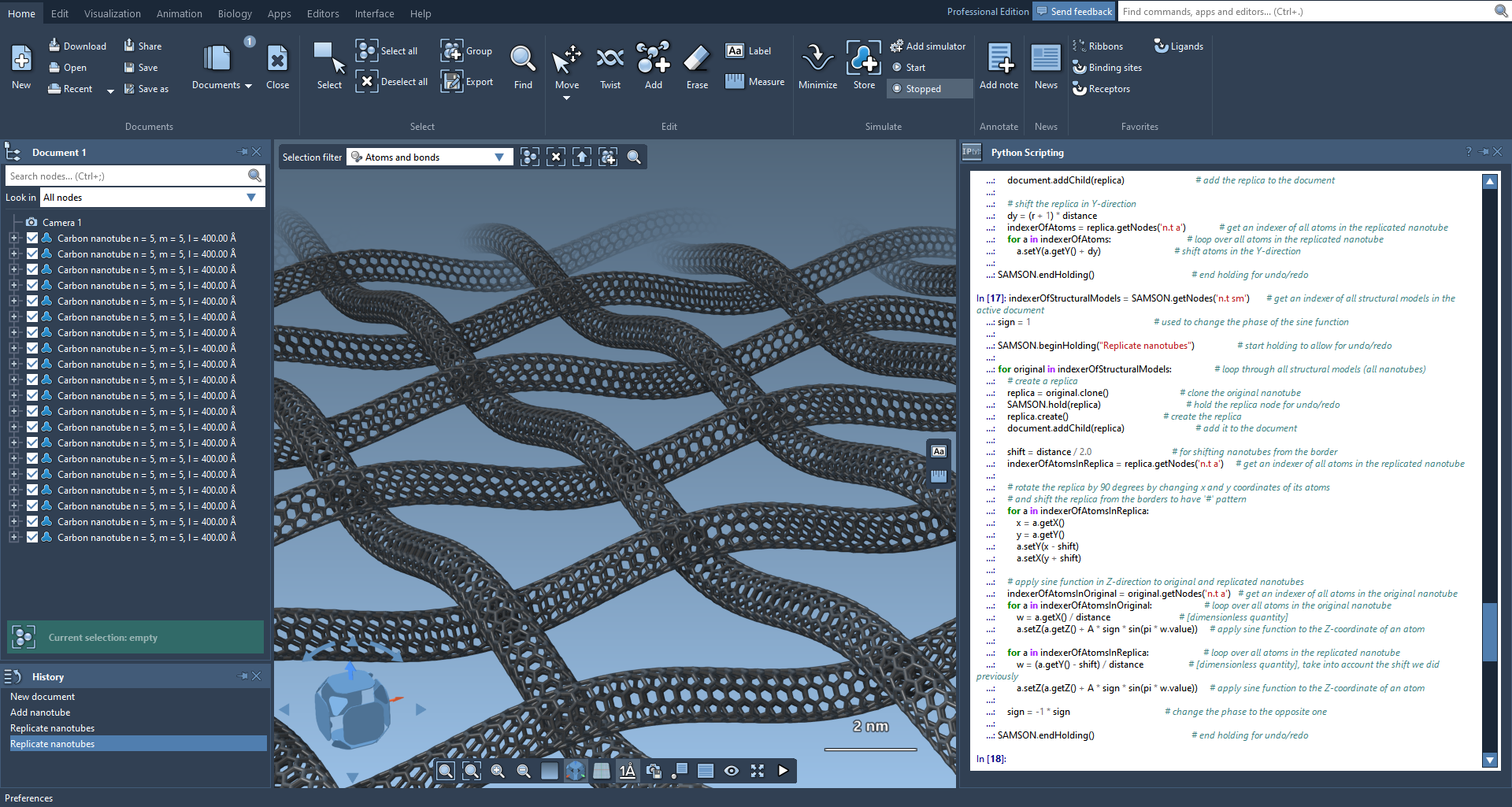
Of course, the created nanotube fabric should be later minimized.
