Select connected components by molecular weight#
This example demonstrates how to choose connected components based on their molecular weight (MW) using a built-in dialog.
Example: Select connected components by molecular weight#
"""
This script provides the possibility to choose connected components based on their molecular weight (MW)
"""
import math
def get_molecular_weight(atom_indexer):
'''
Returns the molecular weight of atoms in the atom_indexer
'''
mass = SBQuantity.mass(0.0)
for atom in atom_indexer:
mass += atom.molecularWeight
return mass.Da
def get_connected_components():
'''
Returns a list of indexers with connected components from the active document
'''
connected_component_list = []
# get all atoms in the active document
atom_indexer = SAMSON.getNodes('node.type atom')
# go through atoms to get their connected components
while len(atom_indexer):
atom = atom_indexer[0]
# get connected component for this atom
connected_component = atom.getConnectedComponent()
connected_component_list.append(connected_component)
# remove atoms of this connected component from the atom indexer
for a in connected_component:
atom_indexer.removeNode(a)
return connected_component_list
connected_component_list = get_connected_components()
masses = []
n_connected_components_per_MW = dict()
for connected_component in connected_component_list:
n_atoms = len(connected_component)
# reduce significant digits in mass
mw = get_molecular_weight(connected_component)
mass_floor = math.floor(mw.value * 1e2) * 1e-2
mass_ceil = math.ceil(mw.value * 1e2) * 1e-2
mass = 0.5 * (mass_floor + mass_ceil)
masses.append(mass)
n_connected_components_per_MW[mass] = n_connected_components_per_MW.get(mass, 0) + 1
# sort in ascending order
n_connected_components_per_MW = dict(sorted(n_connected_components_per_MW.items()))
# print the number of molecules per number of atoms
for key, value in n_connected_components_per_MW.items():
print(f"{value} molecules with MW {key} Da")
# get min and max masses with only 2 significant digits
min_mass = math.floor(min(masses) * 1e2) * 1e-2
max_mass = math.ceil(max(masses) * 1e2) * 1e-2
mass_interval = (min_mass, max_mass)
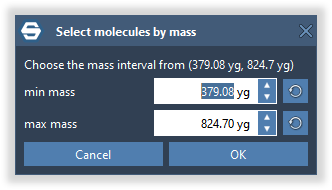
label = f"Choose the mass interval from ({min_mass} Da, {max_mass} Da)"
status, result_mass_interval = SAMSON.getDoubleIntervalFromUser('Select molecules by mass', (label, 'min mass', 'max mass'), mass_interval, mass_interval, mass_interval, (1, 1), '', ' Da')
if status:
# make the operation undoable
with SAMSON.holding("Colorize molecules"):
# clear the current selection
SAMSON.getActiveDocument().clearSelection()
counter = 0
for i, connected_component in enumerate(connected_component_list):
mass = masses[i]
if mass >= result_mass_interval[0] and mass <= result_mass_interval[1]:
counter += 1
for atom in connected_component:
atom.selectionFlag = True
bond_indexer = atom.getBondList()
for bond in bond_indexer:
bond.selectionFlag = True
print(f"{'-' * 40}\nSelected {counter} connected components with mass within ({result_mass_interval[0]} Da, {result_mass_interval[1]} Da)")
else:
# make the operation undoable
with SAMSON.holding("Clear selection"):
# clear the current selection
SAMSON.getActiveDocument().clearSelection()
print(f"{'-' * 40}\nCanceled - clearing the selection")